When Covid-19 appeared towards the back end of 2019, early 2020, most of the planet was focused on how to stop this pathogen spreading. But spread it did, apparently by completely natural means. People who stepped on and off a flight, then a few days later were struck down by this virulent disease, contracted apparently from their point of origin. Remember the first footage of bus loads of passengers being removed to holding facilities for two weeks in an attempt to prevent spread? Do you know one of these passengers from the evacuation flights out of Wuhan?
Fast forward nearly two and a half years and we are pretty certain how these passengers were really contracting Covid-19. Firstly, exposure to arsine before a flight (or possibly during take off through air intake), followed by amplification by lightning induced gamma rays, either in the air or during a storm on the ground. The delay in visible illness being down to the variations in exposure to both elements. The more arsine inhaled and the more air travel, the more likely someone would be to express illness. A very simple, yet invisible, recipe, though of course wildly removed from our 100+ years of established understanding of what a ‘virus’ is.
But of course we still have the elephant in the room. The 30,000 amino acid containing virus and it’s 1273 amino acid long spike protein. The protein was transcribed with speed by the Chinese and contained bits of bats, even HIV and some of what appeared to be human DNA. This inexplicable hotchpotch was claimed later on, by western and Chinese whistleblowers, to be the result of a laboratory engineered virus which could be changed at will and given what is called ‘gain of function’. This is the tweaking of the genetic code to make the virus more effective at it’s job.
What are Exosomes?
A little later, in April 2020, Dr Kaufman started to talk about exosomes, which we thought was a novel idea. As Dr Kaufman’s popularity grew he started doing sessions with other more renown doctors, and his theory seemed to take a back seat to the overarching terrain versus germ theory. If you listen to the video link he mentions arsenic around the 25:30 minute mark and ionising radiation around the 26:10 minute mark as causative factors for exosome expression along with other ‘toxic insults’. Yet talk of 'communicable virus’ continued in practicing medical and scientific circles. The concept of exosomes was ignored, as were the causative factors.
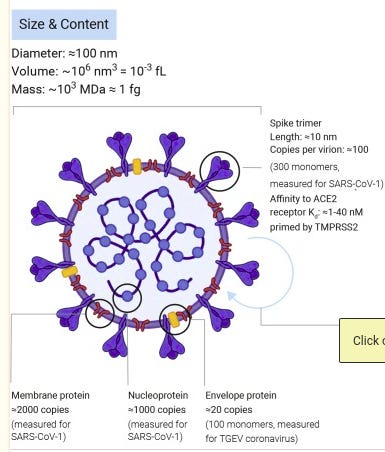
On closer inspection exosomes seemed to be plausible candidates. They are about the right size of 30-100 nanometres. Covid-19 was apparently also around 100 nanometres, or the ‘sneezing’ size of droplet expelled from the body. According to this diagram there were approximately 100 spike proteins per virus and they measured approximately 10 nanometres.
Exosomes are understood to hold peptide markers on their surface proteins that represent the RNA, mRNA, microRNA and other proteins carried in the vesicle. Naturally, peptides bond to the exosome surface proteins as a type of cargo manifest. Experiments have looked at targeting different peptides to exosomes, and importantly also creating chimeric combinations of peptides. They are also present in all bodily fluids including ‘blood, breast milk, saliva, urine, bile, pancreatic juice, cerebrospinal fluid and peritonial fluid’.
This video presentation by Dr Joseph Shehadi tells us a lot more about how exosomes work and how long lasting they are in the system. They can even penetrate the blood brain barrier, help to restructure myelin sheath and even remove amyloid beta particles. These are the proteins that build up in the brains of people with alzheimer’s and other neuro degenerative diseases. Of course science increasingly likes to harness the potential therapeutic benefits of structures produced within the human body. The presentation above deals with stem cell exosomes.
Exosome therapy
This study talks about using stem cells to rebuild the discs of the spinal column, the theoretical effects being wide ranging. They find that exosomes can reduce cell destruction, ‘inflammation, oxidative stress’, early cell death and pro-inflammation cytokines in this study. They are also said to rehyrate the soft centre of the disc, encourage cell and mitochondrial growth and other effects.
Stem cells are fresh new cells that develop in to other cells (differentiate). Taking the exosomes from these should allow the scientist to traffick protein instructions for growth in any number of different of tissue. The thing about exosomes is that they need the right sets of instruction inside them to do the right job. If they carry the wrong instructions they could cause harm to neighbouring cells and overall health.
This article outlines why culturing stem cells in a petri dish is nowhere near as effective as using stem cells themselves together with their exosomes. They liken this to sending a football team out to play a match with no playbook. Exosome therapy is very much still in its infancy it seems.
Exosomes produced by the human body, however, are very much in play and can be produced by almost every cell type. We know that exosomes have the ability to communicate between cells and play a role in reducing illness and disease, but what have they got to do with Covid-19?
What’s inside?
Initially scientists thought these tiny vesicles were just the rubbish collectors of the body. They float around in the cell presumably sucking up bits of dead cells and other ‘stuff’. The cell forms an endosome capsule out of the outer membrane of the cell. Exosomes, formed from organelles, are absorbed in to that endosome. The endosome capsule re-bonds with the membrane of the cell and expells all the exosomes in to the extracellular fluid in the spaces outside of the cells. If they were the dustbins of the cells we would expect them to be engulfed by phagocytes and other immune system cells to remove them from the body.
However, as research has progressed, it appeared that exosomes had a far more exciting role. They could take bits of DNA, mRNA, vRNA, microRNA and other proteins to other cells. They are quite like the gene cassettes that bacteria colonies use. These can send out coded instructions to other cells to produce the right machinery to detoxify heavy metals and other instructions.
As each exosome is different, according to the proteins and coding it is transporting, it isn’t really possible to look at the contents. There would be trillions of possible permutations and we would be here for centuries. Exosomes, however, do tend to communicate certain classes of compounds including enzymes, signalling compounds and chaperones that keep the proteins in the cells functioning well. They also contain proteins that make up the skeleton of the cells (cytoskeleton), interleukins, which activate different immune cells and can promote release of cytokines, antigen presenting-type proteins and of course the proteins that make up the body of the exosomes themselves. All of these can be communicated from cell to cell.
For reference later on. The proteins contained inside exosomes as per the Dr Shehadi video above (1:36 minutes) include: Chaperones - Heat shock proteins 70, 90 and 60 and Heat shock cognate 70, Cytoskeletal proteins – Actin, Tubulin, Cofilin, Profillin, Myocin, Vimentin, Fibronectin, Meosin, Keratin, Talin, Signalling proteins – EGFR, HiF_1alpha, CDC42, P13K, ARF1, Beta Catenin, Enzymes – GAPDH, PK, ATPase, PGK, Enolase and lastly MVBs or multivesicular body forming proteins – Alix, Tsg101, Clatherin, Ubiquitin. The interleukin (IL) group can also be expressed by exosomes. You can look these terms up on www.uniprot.org, which gives a summary of what each protein or gene does.
Exosome surface proteins
Very much like Covid-19 and other ‘viruses’, exosomes use various mechanisms to communicate these proteins and signals to other cells. They have different structures in their two layers (bi-lipid) of membrane. Here are some of them.
Annexins are calcium and lipid binding proteins which are attached to cell and exosome membranes. They behave like stress sensors and have multiple roles in cell survival. Annexins can also be shed by exosomes to other cells as well as being present in the membranes.
Tetraspanins are proteins that span across membranes. They have four domains (sections) with loops either side of the membrane. 33 of these proteins have been identified in humans. Some tetraspanins including CD9 (Tspan29) and CD63 (Tspan30) are said to be present, and play a role, in platelet activation according to the paper ‘Platelet Receptors’ by Kenneth J. Clemetson, Jeannine M. Clemetson. Other tetraspanins can also be present depending on the role of the exosome.
Integrins are the proteins that make the link between the cell and extracellular matrix. They act as receptors for fibronectin, fibrinogen, prothrombin, thrombospondin, E-cadherin and interestingly von Willebrand factor, among other proteins.
Tsg101 or Tumor susceptibility gene (poorly labelled) is essential for normal function of cells. This experiment shows what happens when the gene is taken out of mice. The Alix gene helps deliver the correct tetraspanins to exosomes but also plays a key role in getting the correct miRNA’s in to the exosomes for transport out of the cell. It also plays a role in coding the programmed cell death 6-interacting protein.
Lamp2b is a membrane gene which forms the Lamp2b or lysosomal associated membrane protein 2b. It is used to regulate autophagy in cardiac muscle cells (cardiomyocytes). Autophagy is a crucial part of cell renewal where poorly functioning cells are removed to make way for ones that work better.
MHC I proteins are believed to protect cells from natural killer cells. Any cells that do not express enough are targets. This can happen both with diseased cells and supposed healthy cells. MHC II proteins are found on the ‘surface of antigen-presenting cells display a range of peptides for recognition by the T-cell receptors of CD4+ T helper cells’, but they are also being earmarked as a factor in disease.
Finally we looked at ICAMs. These are intracellular adhesion molecules expressed by immune cells, normal cells and sometimes brain cells. They are all programmed to join with the ‘lymphocyte function-associated antigen-1’ on the surface of white blood cells.
ICAM 1 is apparently also a major rhinovirus (common cold) and Coxsackievirus receptor and is degraded in Kaposi’s Sarcoma herpesvirus. ICAM 3 ties to integrin alpha-D/beta-2, which regulates the death of white blood cells. ICAM 4 ties to alpha-4/beta-1 integrin, which controls cell motility and cell motility receptors, and alpha-V integrin.
Structural comparison
We wanted to look at some of these surface proteins and the Covid-19 spike protein to see if there was anything in the exosome that looked remotely like the Covid-19 protein. To do this we thought of comparing the amino acid codes but that would require cross comparison of multiple sections of code across all of these proteins. A task better suited to a computer algorithm.
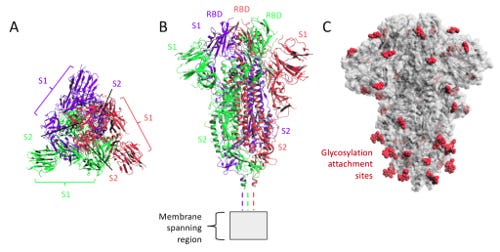
The Covid-19 spike protein has 1273 amino acids. It is made up of a number of sections including a super antigen section that may bind to T-cells, integrin and ACE2 binding areas, and NF9 peptide recognised by CD8+, two cleavage sites (protein chain breaking area), two fusion peptides, a binding site to the trafficking protein that regulates endocytosis, and a binding site to the host membrane.
Like most proteins it contains a signal section which gives instructions for the start of the protein chain and is organised in to two chains S1 and S2, both involved in fusion to the host cell membrane. In the Uniprot database the protein is show to have 15 disulphide bonds, 26 glycosylation sites and 10 lipidation sites. Disulphide bonds maintain the folds in the protein chain. They are also very attractive to arsenic species.
Gylcosylation sites are where proteins or lipids can attach usually aided by enzymes. This can bond the protein to the extracellular matrix and allows the protein to have interactions with the cell. In the spike protein are mostly nitrogen-linked complex, hybrid or high mannose asparagine sites or oxygen-linked threonine sites.
Protein lipidation sites, of s-palmitoyl cysteine, also an arsenic attractive, sulphur containing compound, regulate processes such as ‘membrane trafficking, protein secretion, signal transduction, and apoptosis’.
Exosome structural comparisons
Tetraspanins seem to be much simpler in construction. They are a chain of four sections of tightly coiled protein of varying lengths according to their type and location. They do have glycosylation sites, 1-3 compared to the 26 of the Covid-19 spike protein. They do not have lipidation sites either. Tetraspanin 1 has 3 glycosylation sites and is 241 amino acids long. Visually its tight coils may bear some resemblance to those in the virus membrane-bonded site of the spike in the bottom half of the centreal diagram.
Integrins are longer chains of amino acids and have a number of features similar to the Covid-19 spike. Integrin Alpha 5 (1049 amino acids) has 14 nitrogen-linked asaparagine sites, 9 disulphide bonds and 22 binding sites.This particular integrin has sections of flat amino acid Beta sheets interlaced in to a V shaped arrangement, with a small tightly coiled section coming from one of the sections. Visually it is much simpler than the Covid-19 spike protein but sections may have some similarities with the receptor binding domains (RBD) seen at the host end of the spike (top of diagram).
Lamp2 (b) is 410 amino acids long, consisting of three domains, one in the membrane and one either side of it. Interestingly it is packed tightly with glycosylation sites and is also a glycoprotein. It has nitrogen linked polyactosaminoglyan asparagine (3), normal nitrogen linked asparagine (13), oxygen linked serine (2) and oxygen linked threonine (8) sites. It has no lipidation sites. Visually it has a number of interlaced flat amino acid (Beta) sheets, one tightly coiled region and two loosely coiled regions. It also bears some similarties with the RBD region of the spike protein.
The longest of the ICAM group of proteins, ICAM5 is also positioned inside, outside and through the membrane and consists of 924 amino acids. It has 14 glycosylation sites which are all nitrogen linked asparagine except for one high mannose asparagine site. It also has two modified residues of phosphothreonine where Covid-19 spike protein has none. Visually it is a long series of clumped beta sheets with little resemblance to the spike protein.
All four proteins seem to be composed of similar material, ICAM, Lamp2 (b) and Integrins with similar binding sites to the Covid-19 spike protein. We could not definitively pin the virus spike protein down to one of these proteins, but it could well integrate a number of them and possibly other proteins. A chimeric type spike protein?
Decoding
In an attempt to understand how the spike protein might relate to human exosomes we thought we would try something else. Very unscientific from a molecular biology point of view, but something based on pure logic. To really understand what we were being told was a ‘virus’ it made sense to take a look at the amino acid coding of the spike protein. When we look at proteins they are read left to right from the N-terminal (head) to the C-terminal (tail). Peptide chains are short sections of amino acids anywhere from 2-20 in length also with N- and C-terminals, always joining N- to the previous C- like a long chain of lego bricks. We wondered if we could get from the head to the tail of the spike protein, by finding peptides that could also be present in human proteins. All without breaking the sequence.
The starting M amino acid was left out. All protein chains have them, so this could easily have been added in to make the chain look right. As we could not determine the result of the second, third, fourth etc peptide length before the previous one was finalised, it meant that only one person could do the decode, working one peptide at a time. Putting in the first five letters in sequence into this Japanese protein database, it returned a number of peptides as parts of human proteins. Adding letter by letter to each peptid chain, the returned list was narrowed down to a few or a single human protein entry. If too many letters were added it would return only the Spike protein, perhaps with one mouse protein, or a yeast. Rarely did we return other spike protein codes for other viruses. This in itself was really strange.
It took three days and results were in. It was possible to get to one end of the spike protein to the other, in sequence. The whole chain could be made up of 4, 5, 6, and 7 letter peptides of amino acids, ALL of which could be found in human proteins.
Each protein represented by the peptide being searched was entered in to a log. At the end it was possible to sort it ensuring that each entry had its related peptide against it for checking. The process was done snippet by snippet and there may well be errors, but the overall picture is quite eye opening.
Are there no coincidences?
There were a total of 33 repeats in proteins resulting from this search. As in, there were five entries for Titin (titan protein), two for attractin-like protein 1, two for DNA polymerase theta, three for Fibrocystin-L, two for Lactotransferin and so on.
The most remarkable thing about this decode is that, even where there are repeats like this, the peptides did not match, which means there were more than one peptide section for 33 different proteins in the whole chain. That means also that there were absolutely NO repeats, in 1273 amino acids. We are not experts in probability but we think for there to be not a single repeat in a selection of just 20 letters, in sequentially read peptides, is quite something.
Of course sometimes a protein had various forms, so there may have been a list all appearing with one peptide search. But even when we came across a few more of the same family of proteins in another peptide search the isoforms NEVER repeated. They were all different.
Something else unusual stood out. We could see a number of peptides which would make up part of the proteins contained inside exosomes. We logged ubiquitin related proteins 37 times, keratin appears 13 times, actin 28 times, ATPase 12 times, heat shock protein chaperones and related proteins 9 times, interleukin 7 times, all with different codes and peptide sections, 5 for Rab, 3 for tubulin and 3 for ARF among others.
There were also a staggering 111 entries for receptors of various types including atrial (heart), bone and ephrin. Some renin-angiotensin-system peptides were present, which form part of the blood pressure regulation system, and cytochrome P450 proteins, responsible for liver detoxification. 29 antigen type protein peptides were present. There were even peptides that would appear in proteins governing the sense of smell, 27 were related to DNA and a whopping 53 proteins related to zinc, with the majority being zinc fingers. At least 20 were related to neurons and the central nervous system. All of these areas affected by arsenic, radiation and Covid-19 exposure!
18 peptides formed part of proteins related to the testis and testicular cancer, while other included breast, prostate and skin cancer. Also present were tumour blocking peptides. Looking further there were peptides that would be present in illness such as von Willebrands, HIV, Hepatitis A and Bardet-biedl Syndrome. Remember the rumours of HIV being present? Indeed the structure for one variant of HIV apparently bears similarities to the entire Covid-19 protein.
What are we actually looking at?
As this was a highly irregular method of assessing a protein chain, and we cannot take the results as a fixed template. However, it seems too coincidental that the decode never required reducing the number of amino acids below 4 to get a result, nor was it possible to go over a total of 7 amino acids before results for human proteins completely disappeared, leaving the Spike protein with maybe one or two companions. In some ways, the proteins related to the peptides found are not as important as the rigid adherence to this parameter, and the complete lack of repetition of protein peptides across the entire chain.
Is the Covid-19 spike protein a computer engineered sequence, is that what they meant by it being made in a lab? A computer lab. Have the peptides found in the chain been selected from all humans proteins that are affected by arsenic poisoning and radiation? Did they create an algorithm that could specifically choose a peptide sequence that would represent each area, without repeating? Was the protein code published so that we could decode it in a scientifically unconventional way to lead us closer to the truth?
We can see clearly from experiments targeting peptides to exosome surfaces, that the technology is a long way off being of significant clinical use. In which case, we could probably conclude that this set of peptides is most likely computer generated but not man-made as a ‘virus’. A shopping list of potential targets for the real culprits – arsine gas and lighting induced gamma radiation.
It is also possible the Covid-19 spike protein is based on a REAL surface protein on an exosome, carrying a series of peptides that have been targeted by arsine and radiation. If so, one would expect each exosome cargo and protein to be slightly different. Perhaps this is where the ‘variants’ come in. A little less radiation creates one series of exosome peptides, a little more arsine creates an other series.
Exosomes seem to have facilitated to some degree the development of cancerous tumours and metastisis. Consider how arsenic species readily attach to zinc finger proteins, adjusting coding in the cell. They also disrupt every other cellular function, but prevent cell death by fusing the component that signals a cell to die to the component that prevents cell death. Anything with a disulphide bond is a specific target. Exosome surface proteins are disulphide bond-rich. As many cells still function, so does the endosome/exosome dispatch system, just the cargo they are shipping is more ‘container-like’ and less DHL.
In this article ‘When is a virus an exosome?’ as quoted by Dr Kaufman in the link above, James Hildreth gives a degree of certainty to our theory. ‘“the virus is fully an exosome in every sense of the word.”’ Indeed this article summarises that ‘In the immune system, exosomes transfer peptide-laden MHC proteins to noninfected cells, and also act as miniature versions of antigen-presenting cells.’ Each peptide, from any class with disulphide bonds, as we see from brief list of surface proteins above, is the perfect vehicle by which to spread contamination to other cells.
This healing messenger, the Exosome, has been turned, by an invisible combination of arsine gas and radiation, in to a nanosized time bomb. Multiply that by millions or billions of cells and we have a problem on our hands.
How would you stop it? Create a plug for specific exosome receptors that looks the same but is missing the arsine and radiation?
Thank you to Sebastien as always for your help, insights and particularly for the quote from Hildreth. Thank you Shelly from ENC for the Chinese whistleblower video. Next we will be looking at how arsenic and Covid-19 compare when it comes to the heart.
Cover image is of a 3D printed Covid-19 spike protein courtesy of scitechdaily.com